Tuesday, October 31, 2006
On halloween day Bio2RDF is coming out
We invite you to learn how to write your how rdfizer using the JSTL technology, it is fast and an elegant way to do it. The rdfizer for UniProt, GeneID, Genbank, OMIM, Pubmed, Entrez, GO, ENSEMBL and the PDB are available as GPL program in the /bio2rdf-jsp folder of the application. Read the source, it is a great way to learn for us.
Enjoy and send us your comments at bio2rdf@gmail.com.
Tuesday, September 12, 2006
Try querying Bio2RDF 50 millions triple store with SeRQL
This question illustrates the power of this semantic web technology :
What are the most significant GO terms associated to Paget disease according to UniProt and GeneID annotations using Affymetrix annotations as a crosstable ?
select distinct b1, u1, g1
from
{<http://bio2rdf.org/omim:602080>} <http://bio2rdf.org/bio2rdf#xOMIM> {b},
{b} rdfs:label {b1},
{b} <http://bio2rdf.org/bio2rdf#xOMIM> {c},
{d} <http://bio2rdf.org/affymetrix#xOMIM> {c},
{d} <http://bio2rdf.org/affymetrix#xSwissProt> {u},
{u} rdfs:label {u1},
{u} <http://bio2rdf.org/uniprot#xGO> {g},
{g} rdfs:label {g1}
union
select distinct b1, u1, g1
from
{<http://bio2rdf.org/omim:602080>} <http://bio2rdf.org/bio2rdf#xOMIM> {b},
{b} rdfs:label {b1},
{b} <http://bio2rdf.org/bio2rdf#xOMIM> {c},
{d} <http://bio2rdf.org/affymetrix#xOMIM> {c},
{d} <http://bio2rdf.org/affymetrix#xEntrez_Gene> {u},
{u} rdfs:label {u1},
{u} <http://bio2rdf.org/bio2rdf#xGO> {g},
{g} rdfs:label {g1}
Here is the where to submit it (click on SeRQL-S menu option)
Export result in tabulated format and analyse it with your favorite spreadsheet's pivot table tool.
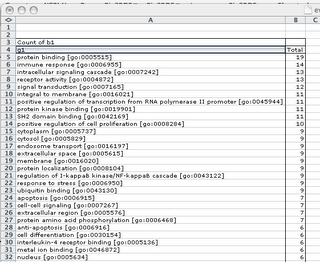 Possibilities are limitless. Enjoy.
Possibilities are limitless. Enjoy.Wednesday, August 30, 2006
Visit Bio2RDF with IBM's lsidBrowser
To install this great Firefox plugin follow the instructions at http://lsid.biopathways.org/lsid_browser/. Then configure the network so that your lsid authority server is http://bio2rdf.org/authority.
Here is what the network configuration should look like :
This is it, you are all set. Try this links it should open in the XUL interactive interface of the lsidBrowser :
lsidres:urn:lsid:bio2rdf.org:mgi:96108
lsidres:urn:lsid:bio2rdf.org:geneid:3131
lsidres:urn:lsid:bio2rdf.org:uniprot:Q8BW74
lsidres:urn:lsid:bio2rdf.org:gi:1484862
lsidres:urn:lsid:bio2rdf.org:genbank:AA021027
lsidres:urn:lsid:bio2rdf.org:go:0048511
lsidres:urn:lsid:bio2rdf.org:pubmed:11641270
lsidres:urn:lsid:bio2rdf.org:omim:142385
Bio2RDF have been launch for testing.
During the conference call of 17 july I informed the members of the HCLSIG BioRDF Subgroup of the availability of Bio2RDF for testing.
Francois Belleau (FB) - Masters in Bioinformatics at Quebec – Focused on applying Semantic Web to build useful knowledge bases. Knowledgebase includes Uniprot, and NCBI data sources. The URL is to be publicly announced shortly, but we can have an early preview (http://bio2rdf.org/). Will have a PhD student for RDF design.